Section: New Results
GridVis: Visualisation of Island-Based Parallel Genetic Algorithms
Participants : Waldo Cancino [correspondant] , Hugo Gilbert, Benjamin Bach, Evelyne Lutton, Pierre Collet.
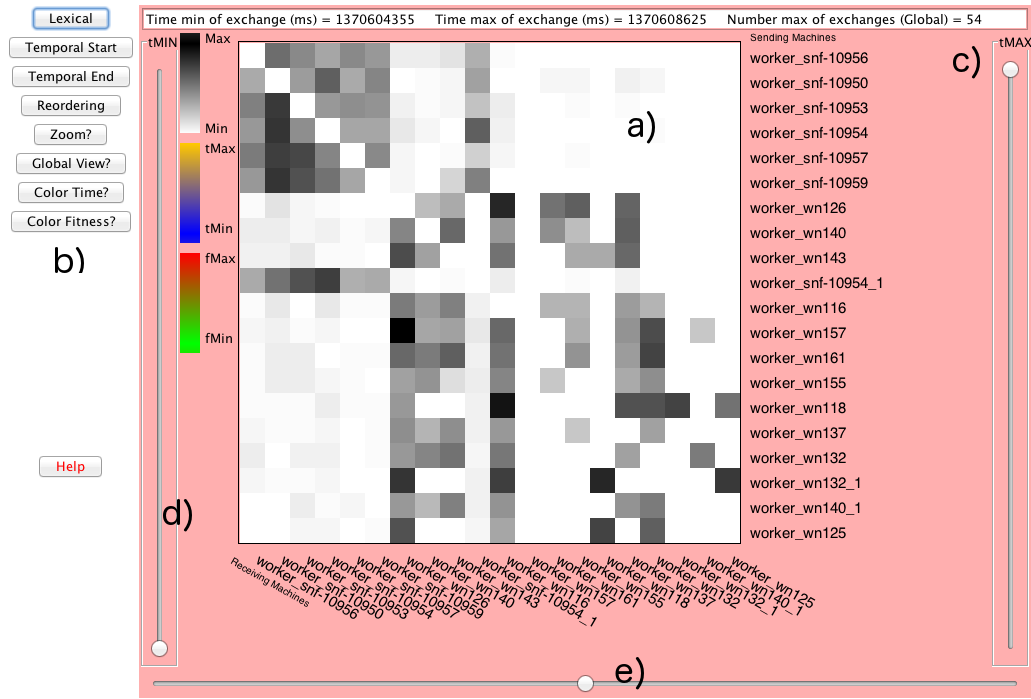
Island Model parallel genetic algorithms rely on various migration models and their associated parameter setting. A fine understanding of how the islands interact and exchange informations is an important issue for the design of efficient algorithms. GridVis, is an interactive tool that has been developed for visualising the exchange of individuals and the propagation of fitness values between islands. GridVis has been developed in Java, to monitor how the islands communicate: when and how much individuals of which fitness they effectively exchange during a run. We model the computer cluster that is running the island model, as dynamic network and use an adjacency matrix to show the relations (exchange between individuals) between nodes (computers) in the cluster (Figure 12 (a)). Several experiments have been performed on a grid and on a cluster to evaluate GridVis' ability to visualise the activity of each machine and the communication flow between machines. Experiments have been made on the optimisation of a Weierstrass function using the EASEA language, with two schemes: a scheme based on uniform islands and another based on specialised islands (Exploitation, Exploration and Storage Islands).
|