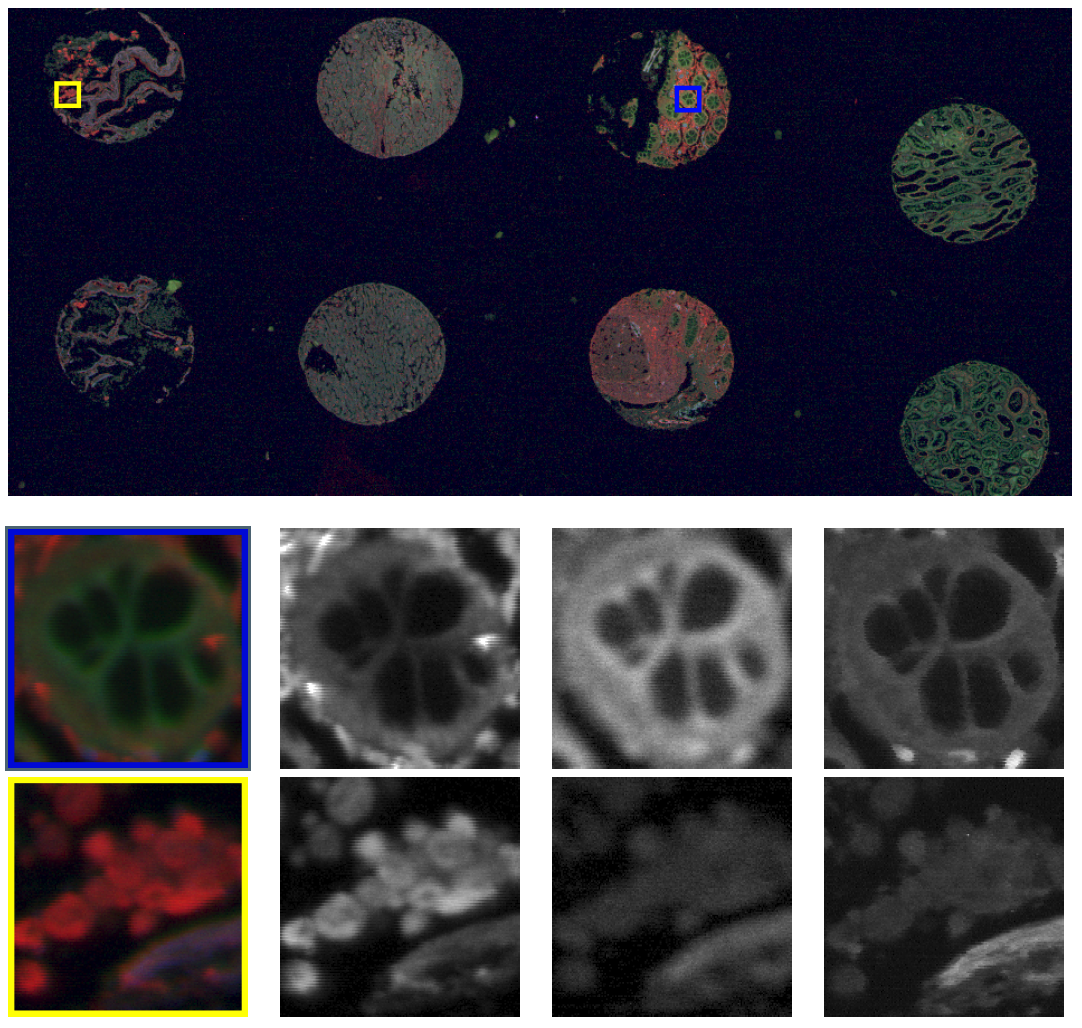
Section: New Results
Algorithms for dejittering and deconvolving fluorescence Tissue MicroArray (TMA) images
Participant : Charles Kervrann.
In the thesis of H.-N. Nguyen, we developed dedicated image processing methods to improve quality of Tissue Microarray (TMA) images acquired by fluorescence scanners. Images are first acquired pixel by pixel along each line, with a change of scan direction between two subsequent lines. Such scanning system often suffers from pixel mis-positioning (jitter) due to imperfect synchronization of mechanical and electronic components. To correct these scanning artifacts, we proposed a variational method based on the estimation of pixel displacements on subsequent lines. This method, inspired from optical flow methods, consists in estimating a dense displacement field by minimizing an energy function composed of a non-convex data fidelity term and a convex regularization term. We used half-quadratic splitting technique to decouple the original problem into two small sub-problems: one is convex and can be solved by standard optimization algorithm, the other is non-convex but can be solved by a complete search. We showed that our method is able to remove efficiently the rolling effect due to jitter, even in the case of huge images and large non-integer displacements.
Second, to improve the resolution of acquired fluorescence images, we introduced a method of image deconvolution by considering a family of convex regularizers. The considered regularizers are generalized from the concept of Sparse Variation which combines the L1 norm and Total Variation (TV) to favors the colocalization of high-intensity pixels and high-magnitude gradient. The experiments showed that the proposed regularization approach produces competitive deconvolution results on fluorescence images, compared to those obtained with other approaches such as TV or the Schatten norm of Hessian matrix. The final deconvolution algorithm has been dedicated to large 2D images acquired with ISO scan imager (see Fig.4). The method is able to process a image in 250 ms (Matlab) with a non optimized implementation.
Collaborators: Vincent Paveau and Cyril Cauchois (Innopys company),
Hoai-Nam Nguyen.
|