Section:
New Results
Robust EEG-based cross-site and cross-protocol classification of states of consciousness
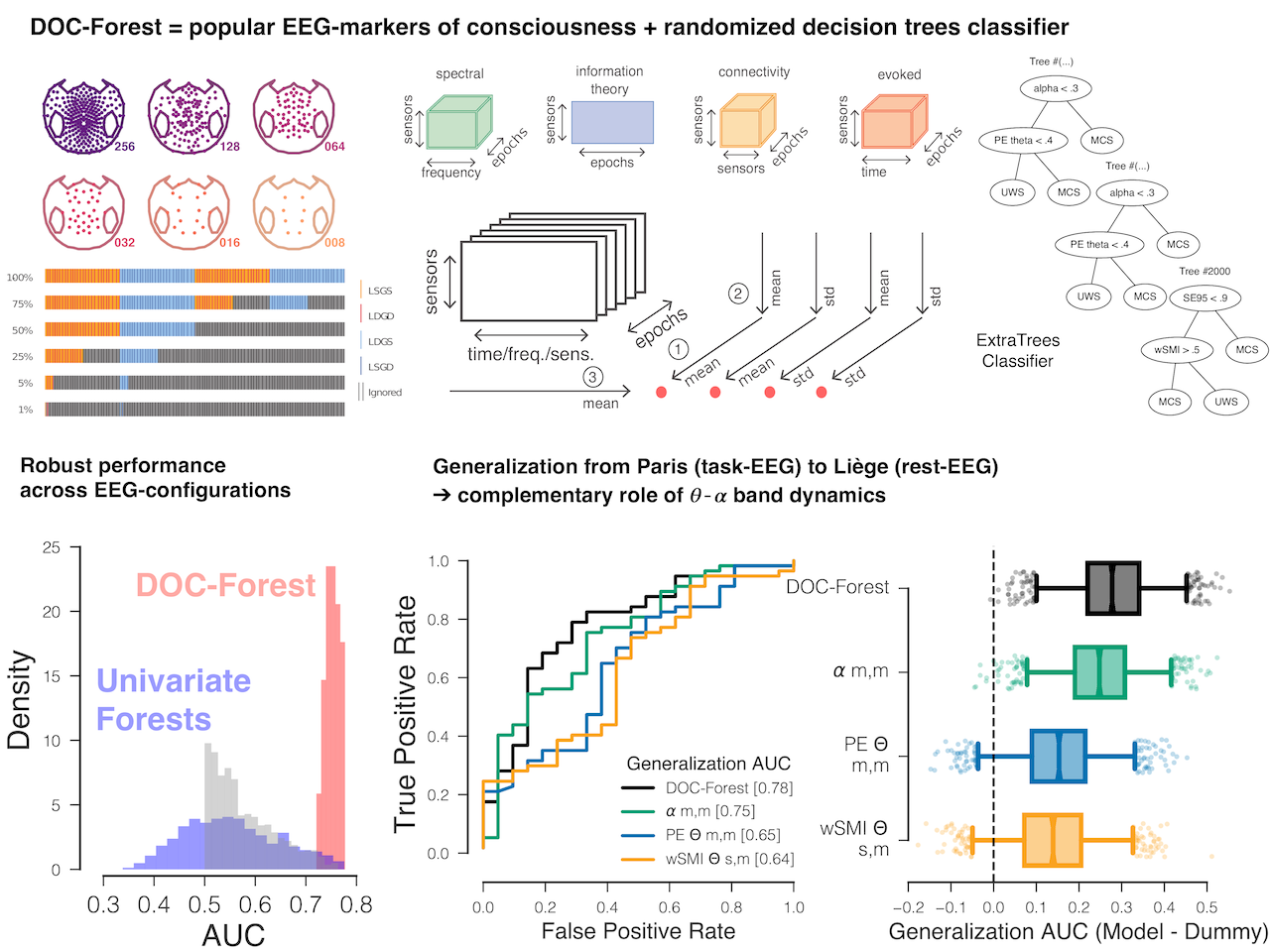
Determining the state-of-consciousness in patients with disorders-of-consciousness (DOC) is a challenging practical and theoretical problem. Recent findings suggest that multiple markers of brain activity extracted from the electroencephalogram (EEG) may index the state of consciousness in the human brain. Furthermore, machine learning has been found to optimize their capacity to discriminate different states of consciousness in clinical practice. However, it is unknown how dependable these EEG-markers are in the face of signal variability due to different EEG-configurations, EEG-protocols and subpopulations from different centers encountered in practice. In our recent paper [11] we addressed the following questions: What is the impact of the EEG configuration (selection of sensors, duration of EEG used)? Do models based on current EEG-markers achieve prospective generalization on independent data from other EEG protocols and other hospitals? Are single markers sufficiently powerful and when does multivariate classification provide the clearest advantage? For summary of methods and approahc see Figure4.
Our results highlight the effectiveness of classical well-studied EEG-signatures such as alpha [8-12Hz] and theta [5-7Hz] frequency band oscillations for detecting consciousness when combined with machine learning. While univariate predictive models achieved good performance, multivariate models showed better generalization capacity and increased robustness to different types of noise while mitigating the impact of the EEG-configuration. Our findings suggest that pooling data over multiple centers for predictive modeling of DOC is a concrete possibility and can become a promising alley for the field of cognitive neurology.
Figure
4. We probed the robustness and validity of EEG-markers of consciousness. Using the robust Extra-Trees algorithm (Geurts, Ernst, & Wehenkel, 2006) we developed a classifier trained to differentiate UWS from MCS patients. This classifier (named “DOC-forest”) was trained and tested using 28 potential EEG-markers of consciousness (112 features) from 249 patients recorded at the Paris Pitié-Salpêtrière and 78 patients from the University Hospital of Liège. We used the MNE-Python software for EEG processing and the scikit-learn package for machine learning. Our results show that optimally combining multiple EEG-markers of states of consciousness using machine learning enables robust generalization across EEG-configurations, EEG-protocols and sites.
Our recipe for extracting biomarkers is available on Github: https://nice-tools.github.io/nice. For a neuroscientific discussion of our work see the accompanying commentary article by Sokoliuk and Cruse (https://doi.org/10.1093/brain/awy267).
|
|