Section: New Results
Geometric view of biodiversity and molecular taxonomy
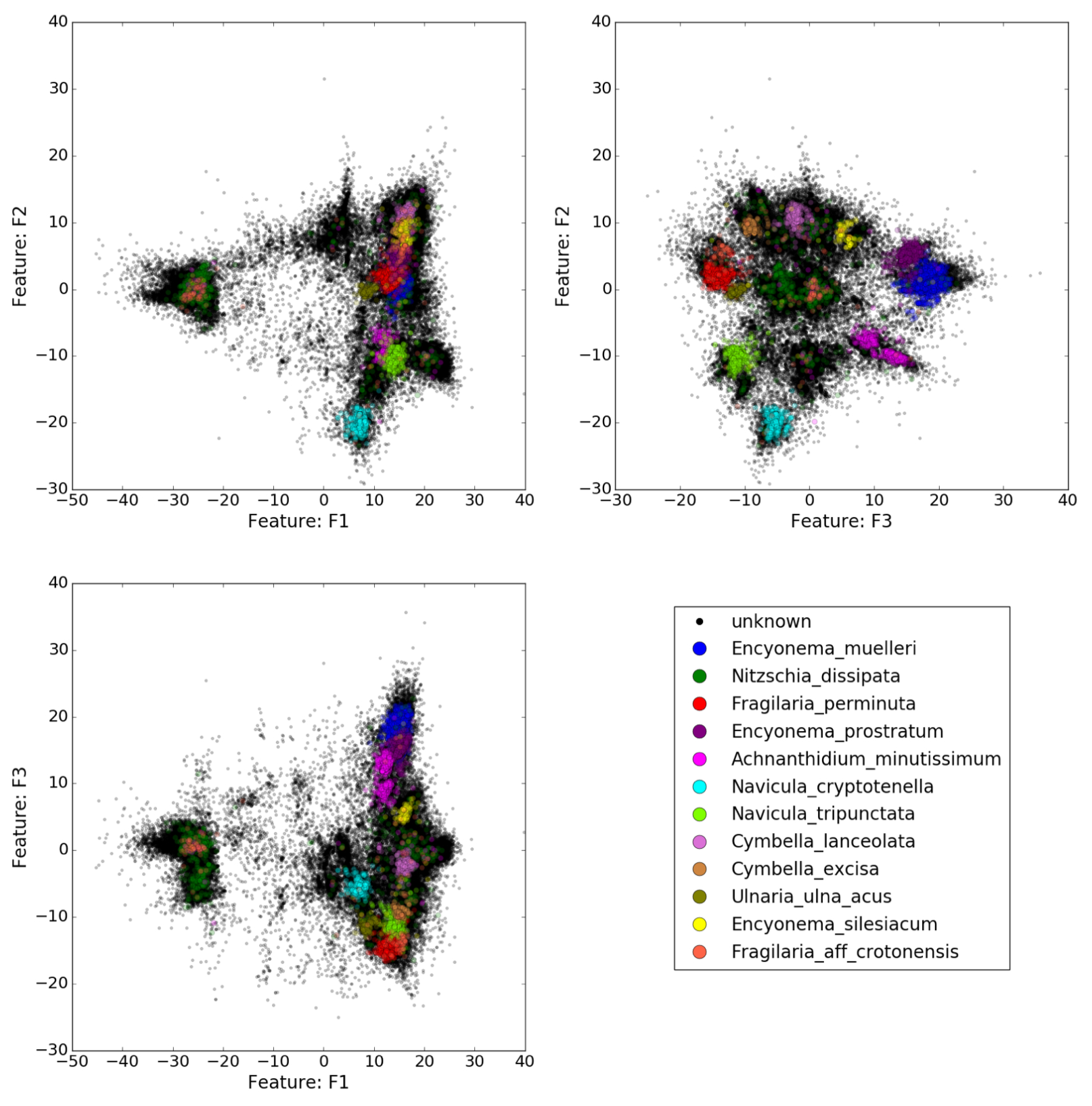
For the geometric view on biodiversity and molecular based taxonomy (metabarcoding), 2018 has been characterized by significant progress in the connections between the questions posed by metabarcoding, and the implementaton of solutions by intensive computing in collaboration with HiePACS team. We are now able to run a Multidimensional Scaling (MDS) to build a point cloud where each point is a read (a short sequence from an environmental sample) on a sample of nearly reads on one node of a cluster with shared memory. Pleiade is member of two projects lead by Inria BSO to extend these possibilities to work with distributed memory (connection with the Chameleon dense linear algebra solver) with the possibility to reach results with one million sequences at hand. To our knowledge, such a connection between scientific questions related to biodiversity and HPC-HTC has up to now had no equivalent and is new.
In collaboration with HiePACS, the SED of Inria BSO, and the GRICAD (Mésocentre of Grenoble), Pleiade has developed a scientific library of tools for dimension reductions in the framework of Big Data, specifically linear algebra of full matrices. A prototype was developed in numpy, then reimplemented in C++ with an external library for Random Projection, and made available in Julia. This library will be connected with the workflow developed in Regional project and ADT Gordon.