Section: New Results
Tangible exploration of protein families
Protein families[3] are an effective way to compare the complete genomes of fungal species. In general these comparisons are very challanging due to the large evolutionary distances involved, the wide range of GC compositions observed from one species to the next, and the extensive map reshuffling that is characteristic of the yeasts in particular. Protein families are a classification of protein-coding gene sequences into phylogenetic groups, using clustering methods and semi-supervised classification. Members of a family are homologous and in many cases this homology is suggestive of functional similarity.
|
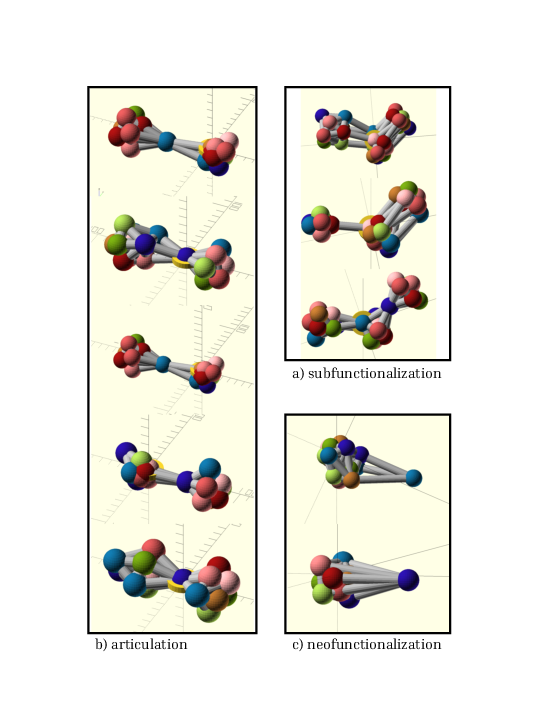
An intriguing feature of protein families is that the weighted graph constructed from their pairwise distance matrices has a structure that reflects the evolutionary history of the family. We developed software (family-3d, see above) that uses truncated distances to construct a weighted graph and to lay it out using an adaptation of the three-dimensional extension of the Kamada-Kawai force-directed layout. The resulting shapes for a set of protein families are then clustered manually by similarity (figure 5). Similarity in shape is highly suggestive of similarity in evolutionary scenarios, leading to hypotheses about the histories of individual protein families and the mechanisms by which functional diversity is obtained.