Section: New Results
Computational Cardiology & Image-Based Cardiac Interventions
Cardial Electrophysiological Model Learning and Personalisation
Participants : Nicolas Cedilnik [Correspondant] , Ibrahim Ayed [Sorbonne, LIP6, Paris] , Hubert Cochet [IHU Liryc, Bordeaux] , Patrick Gallinari [Sorbonne, LIP6, Paris] , Maxime Sermesant.
This work is funded by the IHU Liryc, Bordeaux.
modelling, electrophysiology, ventricular tachycardia, ischemic cardiomyopathy
This project aims at making electrophysiological model personalisation enter clinical practice in interventional cardiology. During this year:
-
we evaluated a fully automated computed tomography-based model personalisation framework in the context of post-ischemic ventricular tachycardia [35],
-
we developped a model personalisation methodology based on invasive data in our participation in the STACOM2019 modelling challenge [37],
-
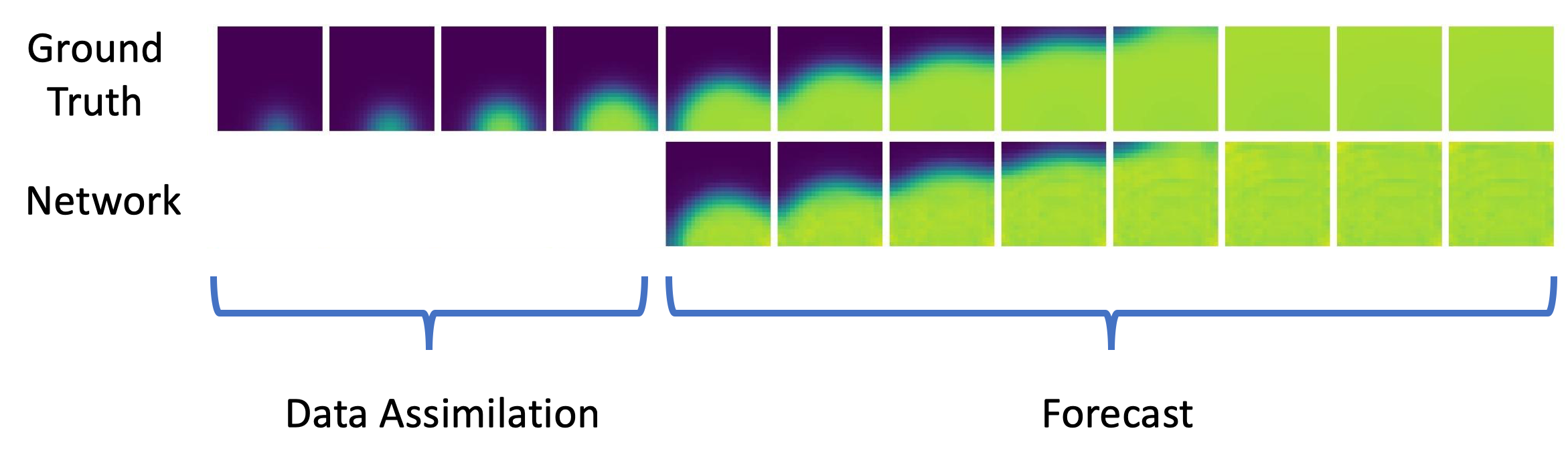
we proposed a deep learning based approach to replace numerical integration of partial differential equations used in cardiac modelling [32], see Figure 17.
|
Deep Learning Formulation of ECGI for Data-driven Integration of Spatiotemporal Correlations and Imaging Information
Participants : Tania Marina Bacoyannis [Correspondant] , Hubert Cochet [IHU Liryc, Bordeaux] , Maxime Sermesant.
This work is funded within the ERC Project ECSTATIC with the IHU Liryc, in Bordeaux.
Deep Learning, Electrocardiographic Imaging, Inverse problem of ECG, Electrical simulation, Generative Model.
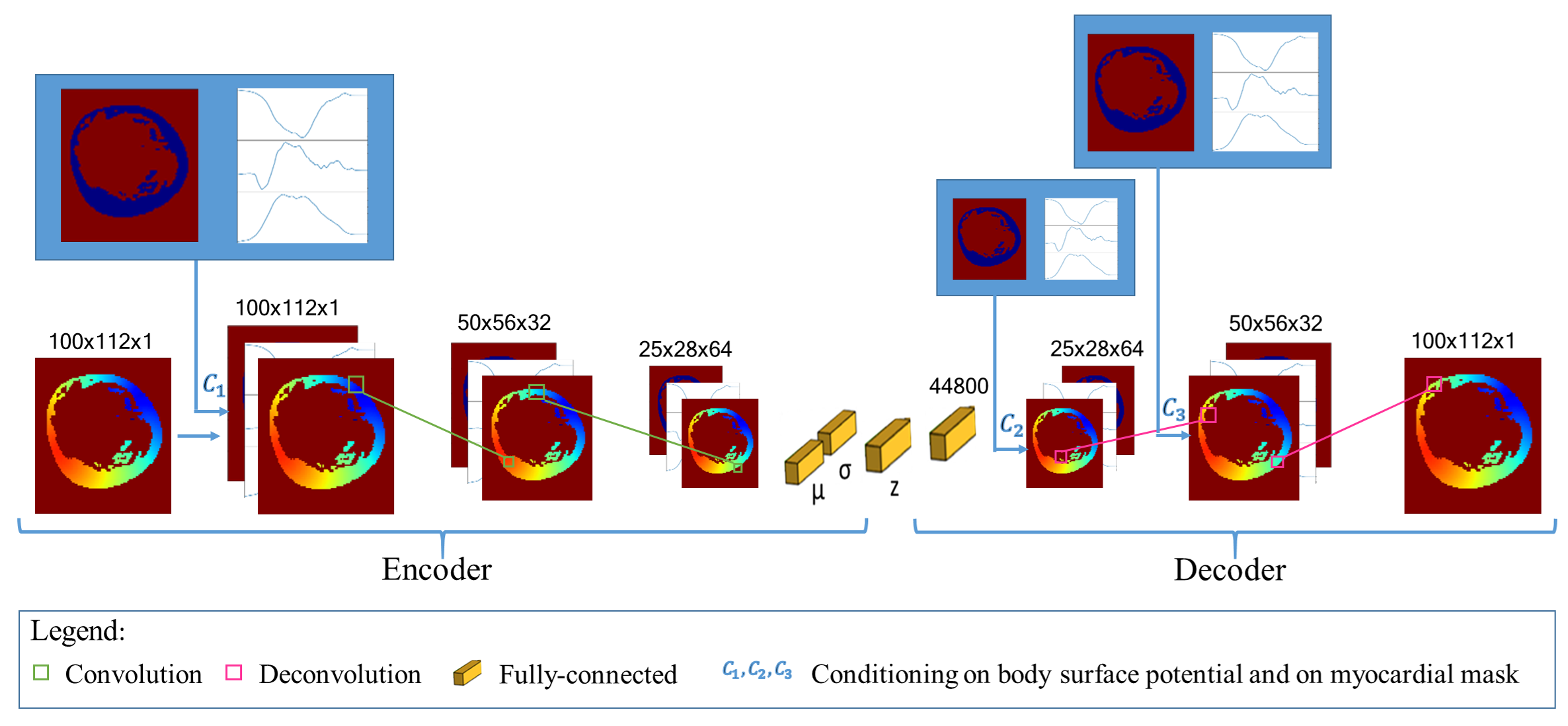
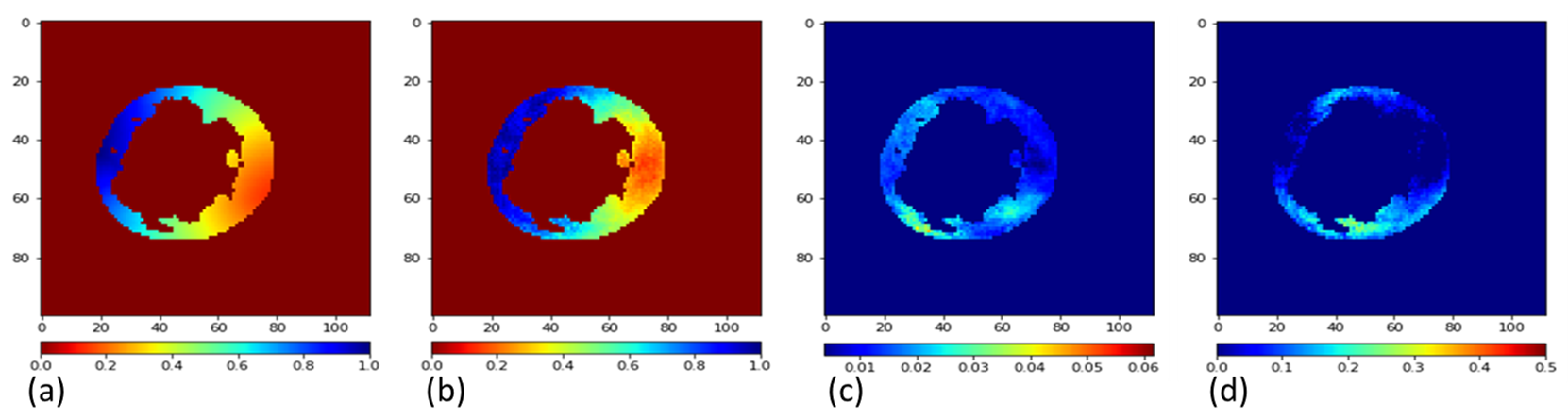
Electrocardiographic imaging (ECGI) aims at reconstructing the electrical activity of the heart using body surface potentials.To achieve this one has to solve the ill-posed inverse problem of the torso propagation. We propose in [33] a novel Deep Learning method based on Conditional Variational Autoencoder able to solve ECGI inverse problem in 2D. This generative probabilistic model learns geometrical and spatio-temporal information and enables to generate the corresponding activation map of the specific heart.
120 activation maps and the corresponding Body Surface Potentials (BSP) were generated using the dipole formulation. 80% of the simulated data was used for training and 20% for testing.We generate 10 propbable solutions for each given input using our model. The Mean Squarre Error (MSE) metric over all the tests was 0.095. As results we were able to observe that the reconstruction performs well. Next, we will extend the model in 3D and test it on real data provided by the IHU Liryc.
|
|
Discovering the link between cardiovascular pathologies and neurodegeneration through biophysical and statistical models of cardiac and brain images
Participants : Jaume Banus Cobo [Correspondant] , Marco Lorenzi, Maxime Sermesant.
Université Côte d'Azur (UCA)
Lumped models - Biophysical simulation - Statistical learning
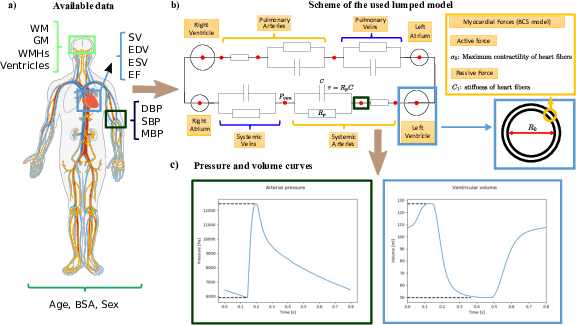
The project aims at developing a computational model of the relationship between cardiac function and brain damage from large‐scale clinical databases of multi‐modal and multi‐organ medical images. The model is based on advanced statistical learning tools for discovering relevant imaging features related to cardiac dysfunction and brain damage; these features are combined within a unified mechanistic framework to providing a novel understanding of the relationship between cardiac function, vascular pathology and brain damage. [34]
|
Parallel transport of surface deformations from pole ladder to symmetrical extension
Participants : Shuman Jia [Correspondant] , Nicolas Guigui, Nicolas Duchateau, Pamela Moceri, Maxime Sermesant, Xavier Pennec.
The authors acknowledge the partial funding by the Agence Nationale de la Recherche (ANR)/ERA CoSysMedSysAFib and ANR MIGAT projects.
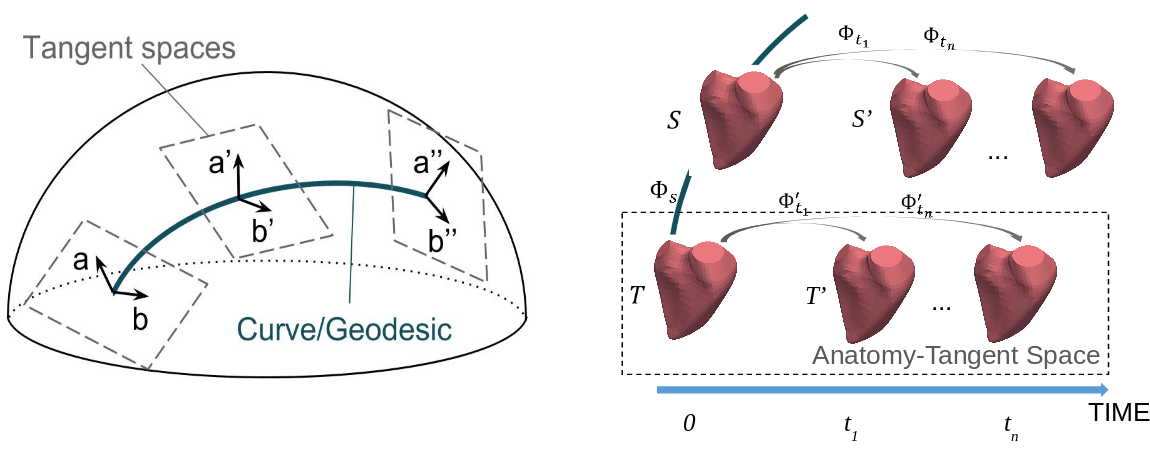
We proposed a general scheme to perform statistical modeling of the temporal deformation of the heart, directly based on meshes. We encoded the motion and the intersubject shape variations, with diffeomorphisms parameterized either by stationary SVFs or by time-varying velocity fields in the LDDMM framework.
Experiments on a 4D right-ventricular endocardial meshes database demonstrated the stability of our transport algorithm, of importance for the assessment of pathological changes. The method is adaptable to other anatomies with temporal or longitudinal data.
|
Machine Learning and Pulmonary hypertension
Participants : Yingyu Yang [Correspondant] , Stephane Gillon, Jaume Banus Cobo, Pamela Moceri, Maxime Sermesant.
cardiac modelling, machine learning
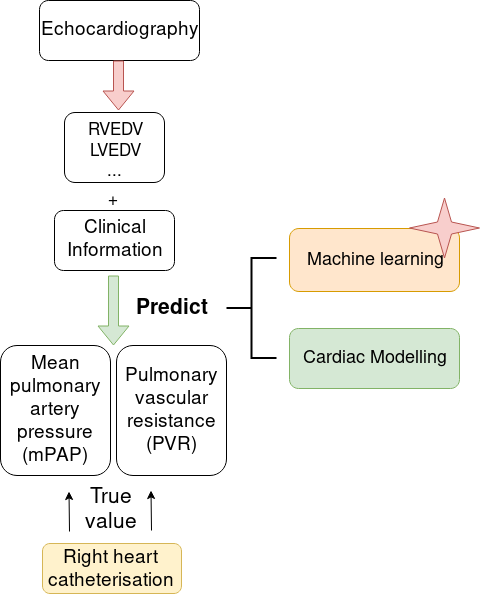
Right heart catheterisation is considered as the gold standard for the assessment of patients with suspected pulmonary hyper-tension. It provides clinicians with meaningful data, such as pulmonary capillary wedge pressure and pulmonary vascular resistance, however its usage is limited due to its invasive nature. Non-invasive alternatives, like Doppler echocardiography could present insightful measurements of right heart but lack detailed information related to pulmonary vasculature. In order to explore non-invasive means, we studied a dataset of 95 pulmonary hypertension patients, which includes measurements from echocardiography and from right-heart catheterisation. We used data extracted from echocardiography to conduct cardiac circulation model personalisation and tested its prediction power of catheter data. Standard machine learning methods were also investigated for pulmonary artery pressure prediction. Our preliminary results demonstrated the potential prediction power of both data-driven and model-based approaches. It was published as "Non-Invasive Pressure Estimation in Patients with Pulmonary Arterial Hypertension: Data-driven or Model-based?" accepted at 10th Workshop on Statistical Atlases and Computational Modelling of the Heart, Oct 2019, Shenzhen, China [46]
Style Data Augmentation for Robust Segmentation of Multi-Modality Cardiac MRI
Participants : Buntheng Ly [Correspondent] , Hubert Cochet [IHU Liryc, Bordeaux] , Maxime Sermesant.
Image Segmentation. Multi-modality, Cardiac Magnetic Resonance Imaging, Late Gadolinium Enhanced, Deep Learning
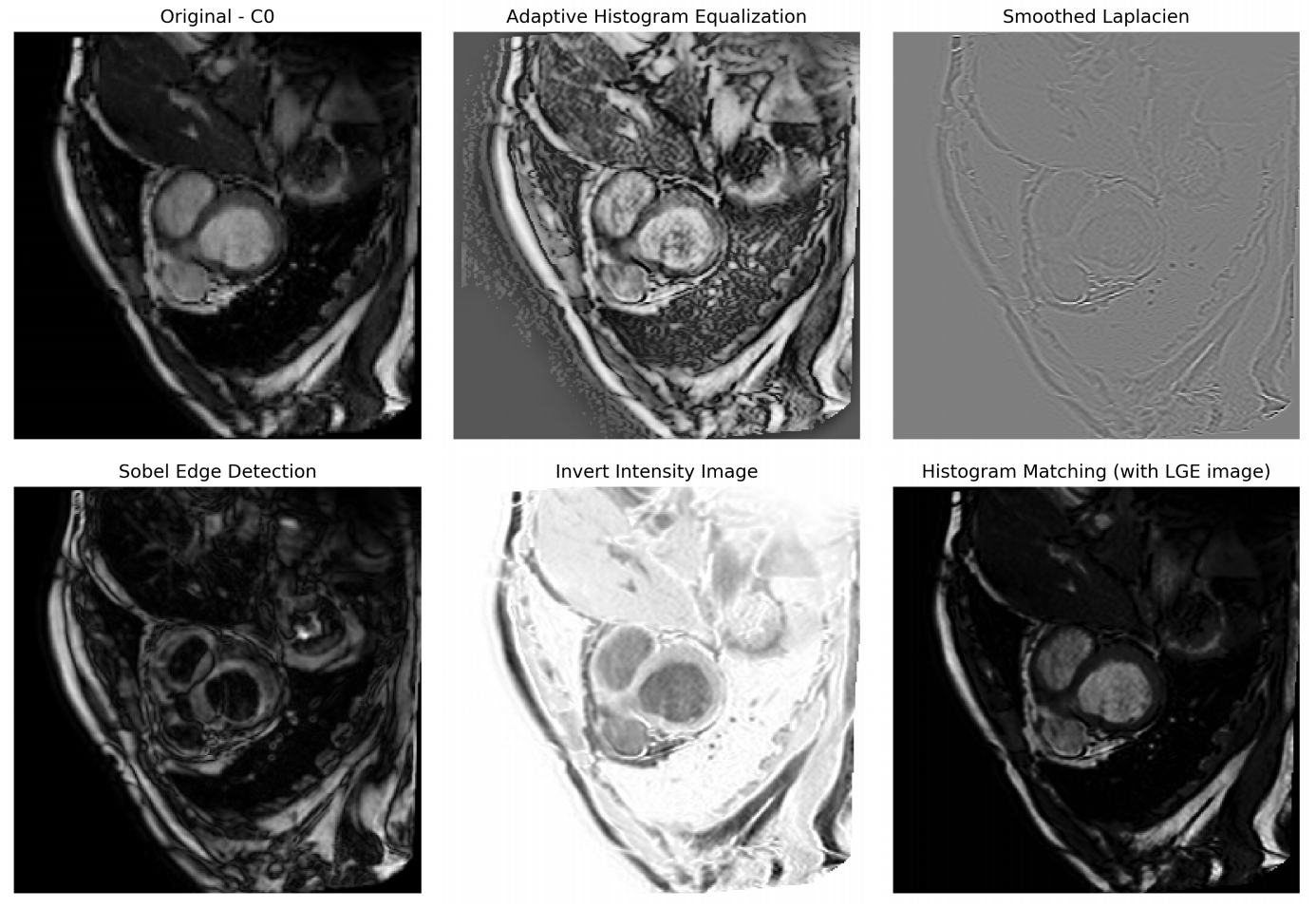
We propose a data augmentation method to improve the segmentation accuracy of the convolutional neural network on multi-modality cardiac magnetic resonance dataset [41].
The strategy aims to reduce over-fitting of the network toward any specific intensity or contrast of the training images by introducing diversity in these two aspects, as shown in figure 23.
|
Towards Hyper-Reduction of Cardiac Models using Poly-Affine Deformation
Participants : Gaëtan Desrues [Correspondant] , Hervé Delingette, Maxime Sermesant.
Model Order Reduction, Finite Elements Method, Affine Transformation, Meshless
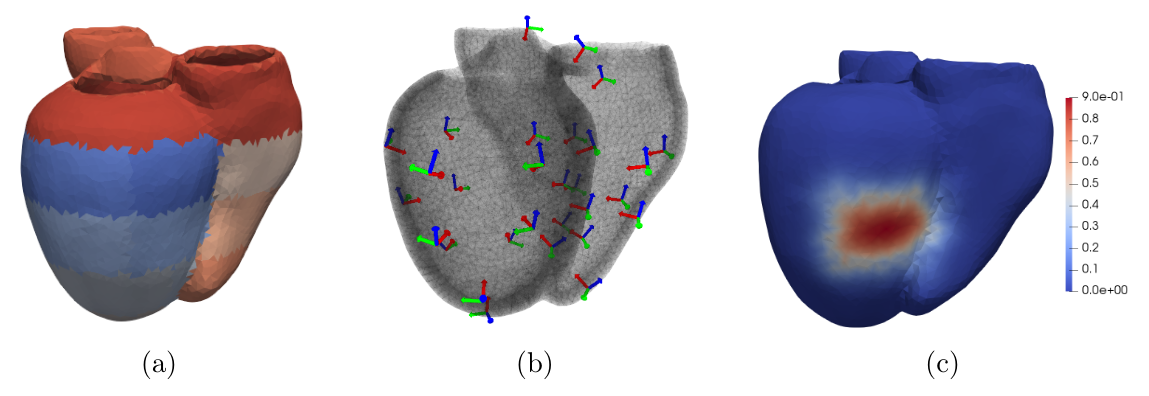
Patient-specific 3D models can help in improving therapy selection, treatment optimization and interventional training. However, these simulations generally have an important computational cost. The aim of this project is to optimize a 3D electromechanical model of the heart for faster simulations [38]. The cardiac deformation is approached by a reduced number of degrees of freedom represented by affine transformations (frames in Figure 24b) located at the center of the AHA regions (Figure 24a). The displacement of the material points are computed using region-based shape functions (Figure 24c).