Section: New Software and Platforms
Platforms
Mobyle@serpico platform and software distribution
Participants : Sylvain Prigent, Léo Maury, Charles Kervrann.
The objective is to disseminate the distribution of serpico image processing software in the community of cell biology and cell imaging.
Free binaries: software packages have been compiled for the main operating systems (Linux, MacOS, Windows) using CMake (see http://www.cmake.org/). A few of them are freely available on the team website https://team.inria.fr/serpico/software/ under a proprietary license.
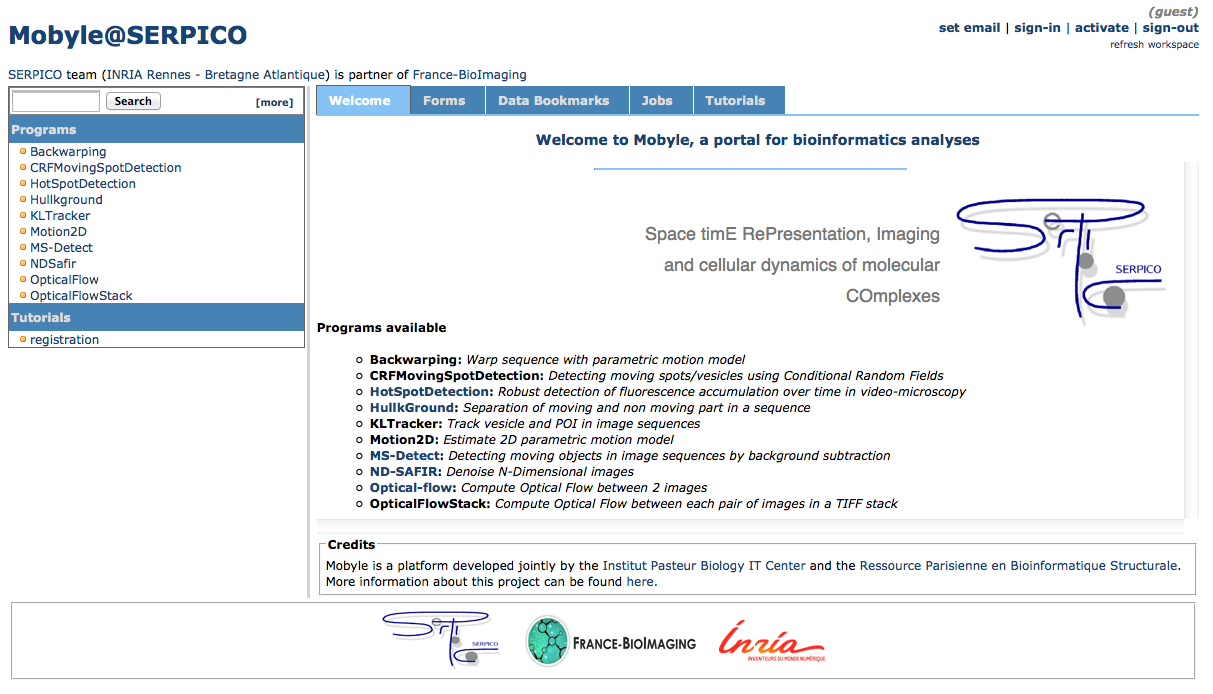
Mobyle@serpico web portal: An on-line version (http://mobyle-serpico.rennes.inria.fr) of the image processing algorithms has been developed using the Mobyle framework (Institut Pasteur, see http://mobyle.pasteur.fr/). The main role of this web portal (see Fig. 2) is to demonstrate the performance of the programs developed by the team: QuantEv , C-Craft [9], Atlas [1], Hullkground [48], KLTracker [52], Motion2D [51], MS-detect [49], ND-Safir [4], OpticalFlow and Flux Estimation [9]. The web interface makes our image processing methods available for biologists at Mobyle@serpico without any installation or configuration on their own. The size of submitted images is limited to 200 MegaBytes per user and all the results are kept 15 days. The web portal and calculations run on a server with 2 CPU x 8 cores, 64 GigaBytes of RAM (500 MegaBytes for each user. Data is saved for 3 months).
ImageJ plugins: ImageJ (see http://rsb.info.nih.gov/ij/) is a widely used image visualization and analysis software for biologist users. We have developed ImageJ plug-in Java versions of the following software: ND-Safir [4], Hullkground [48], Motion2D [51], Atlas [1]. The C-Craft [9], QuantEv and GcoPS [19] algorithms have been developed for the image processing ICY platform (http://icy.bioimageanalysis.org/).
Partners: CNRS-UMR 144 Institut Curie & France-BioImaging (UMS 3714 CEMIBIO).
Bioimage-IT for bioimage management and processing
Participants : Sylvain Prigent, Cesar Augusto Valades Cruz, Léo Maury, Jean Salamero, Charles Kervrann.
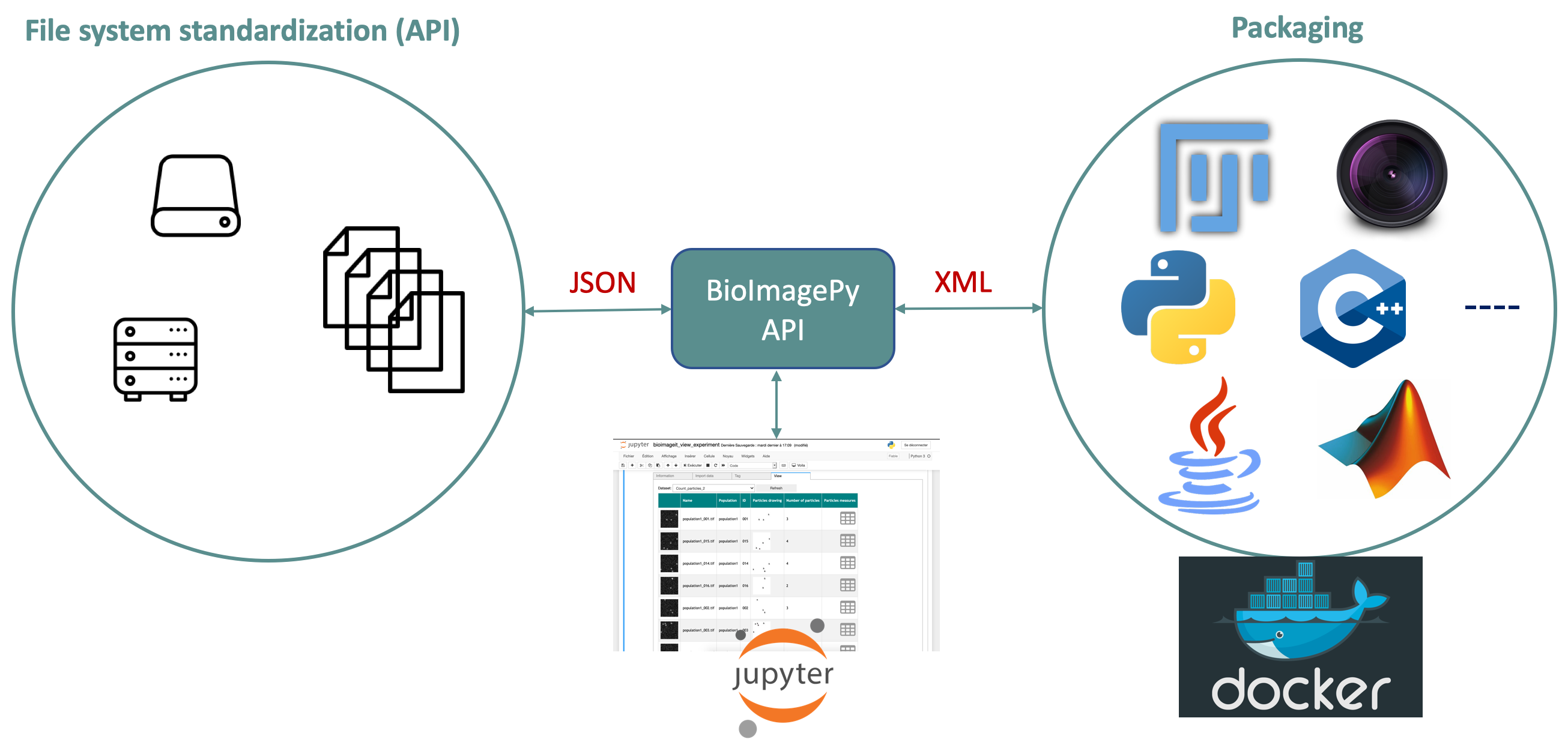
New image acquisition systems generate large number of images and large volume images. Such data sets are hard to store, to process and to analyze for one user in a workstation. Many solutions exist for data management (e.g. Omero, OpenImadis), image analysis (e.g. Fiji, Icy, CellProfiler) and statistics (e.g R). Each of them has its specificities and several bridges have been developed between pieces of software. Nevertheless, in many use-cases, we need to perform analysis using tools that are available in different pieces of software and different languages. It is then tedious to create a workflow that brings the data from one tool to another. It needs programing skills and most of the time, a dedicated script using a dedicated file system for processed data management is developed. The aim of BioImage-IT is to create a “bandmaster” application that allow any scientist to annotate, process, and analyze data using only one single high level application. This BioImage-IT application is based on 3 components:
-
an image processing and analysis tools integration method based on Docker and XML commands description,
-
an application with a graphical interface to easily annotate data, run processing tools, and visualize data and results.
This software architecture has three main goals. First, data are annotated using a file system. This means that data are not dependent on any software like a SQL database, and each experiment can then be stored in a different directory and can be moved from one server to another or to any drive with a simple copy pasting operation. Second, the processing tools are used as binary packages managed by the Docker technology. Docker enables to gently handle dependencies and several versions of the same tool. Any existing tool can then be integrated in its native programming language. Third, using a single “bandmaster” application allows one to automatically generate metadata for any processed data, improving the traceability and the repeatability of any experimental result.
BioImage-IT (https://project.inria.fr/bioimageit) is developed in the context of the France-BioImaging research infrastructure in coordination with the IPDM-FBI (Image Processing and Data Management) node in order to provide a standardized image processing tool set and data management for the imaging facilities.
Partners: CNRS-UMR 144 and U1143 INSERM/CNRS-UMR 3666, Institut Curie & France-BioImaging (UMS 3714 CEMIBIO).