Section: New Results
Dense mapping of intracellular diffusion and drift from single-particle tracking data analysis
Participants : Antoine Salomon, Cesar Augusto Valades Cruz, Charles Kervrann.
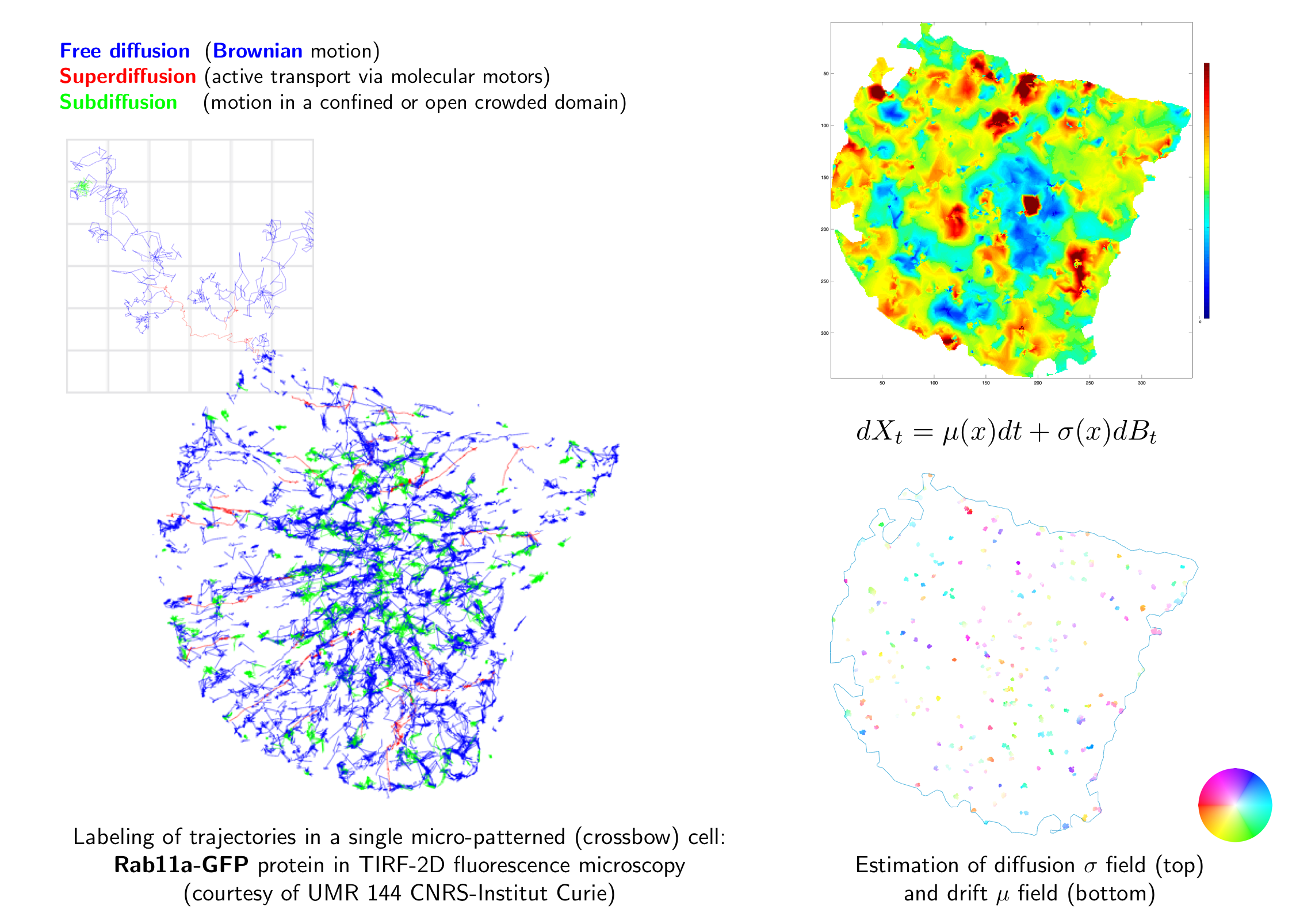
It is of primary interest for biologists to be able to locally estimate diffusion and drift inside a cell. In our framework, we assumed that particle motion is governed by the following Langevin equation: where denotes the 2D or 3D spatial coordinates of the particle, the drift vector, the diffusion coefficient, and the standard Gaussian white noise. In that context, we proposed a new mapping method inspired from [50] that developed a method providing results in the form of matrices by scanning the data by blocks, and from the framework in [5], dedicated to both classifying particle motion types and detecting potential motion switches along a trajectory. To avoid the calculation of both drift and diffusion in cell coordinates where no data is available, we replaced the scanning movement of an averaging window by a Gaussian window centered on trajectory points. Each drift vector and each diffusion coefficient are calculated at coordinates corresponding exactly to the coordinates given by the preliminary particle tracking, which provides more details. A nonparametric three-decision test enables to label trajectories or sub-trajectories [5]. This information is then used. to calculate drift and diffusion coefficient (or Kramers-Moyal coefficients) maps separately on each class of motion with the most appropriate diffusion models: confined motion (the particle is bound to a specific point), Brownian motion (the particle moves randomly), and directed motion (the particle moves in a given direction) (see Figure 5).
Software: THOTH and CPAnalysis (see Sections 6.2 and 6.6).
Collaborators: V. Briane (UNSW Sydney, School of Medical Sciences, Australia),
L. Leconte and J. Salamero (CNRS-UMR 144, Institut Curie, PSL Research University),
L. Johannes (U1143 INSERM / CNRS-UMR 3666, Institut Curie, PSL Research University),
E. Derivery (MRC laboratory of Molecular Biology, Cambridge, UK),
L. Muresan (Cambridge Advanced Imaging Centre, UK).