Section: Software
Software for live cell imaging
Participants : Charles Kervrann, Patrick Bouthemy, Tristan Lecorgne.
Motion2d: parametric motion model estimation
The Motion2D software written in c ++ (APP deposit number: FR.001.520021.001.S.A.1998.000.21000 / release 1.3.11, January 2005) and java (plug-in ImageJ (http://rsbweb.nih.gov/ij/ ) is a multi-platform object-oriented library to estimate 2D parametric motion models in an image sequence. It can handle several types of motion models, namely, constant (translation), affine, and quadratic models. Moreover, it includes the possibility of accounting for a global variation of illumination. The use of such motion models has been proved adequate and efficient for solving problems such as optic flow computation, motion segmentation, detection of independent moving objects, object tracking, or camera motion estimation, and in numerous application domains (video surveillance, visual servoing for robots, video coding, video indexing), including biological imaging (image stack registration, motion compensation in videomicroscopy). Motion2D is an extended and optimized implementation of the robust, multi-resolution and incremental estimation method (exploiting only the spatio-temporal derivatives of the image intensity function). Real-time processing is achievable for motion models involving up to six parameters. Motion2D can be applied to the entire image or to any pre-defined window or region in the image.
Free academic software distribution: Motion2D Free Edition is the version of Motion2D available for development of Free and Open Source software only. More information on Motion2D can be found at http://www.irisa.fr/vista/Motion2D and the software can be downloaded at the same Web address.
On-line demo: Mobyle@GenOuest Bioinformatics http://mobyle.genouest.org/cgi-bin/Mobyle/portal.py#forms:Motion2D (see Fig. 5 ).
Partner: Fabien Spindler (Inria Lagadic team).
ND-Safir and Fast2D-SAFIR: Image denoising software
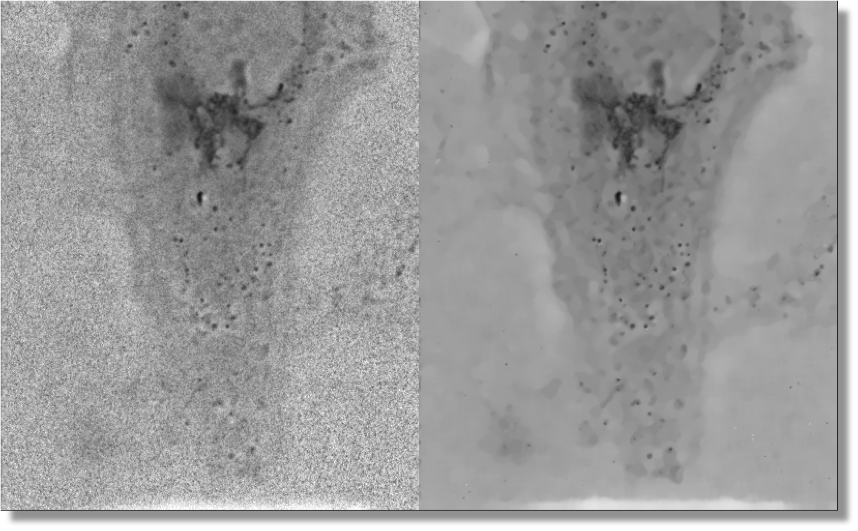
The nD-Safir software (APP deposit number: IDDN.FR.001.190033.002.S.A.2007.000.21000 / new release 3.0 in 2013) written in c ++, java and matlab , removes additive Gaussian and non-Gaussian noise in still 2D or 3D images or in 2D or 3D image sequences (with no motion computation) (see Figure 2 ) [4] . The method is unsupervised and is based on a pointwise selection of small image patches of fixed size (a data-driven adapted way) in spatial or space-time neighbourhood of each pixel (or voxel). The main idea is to modify each pixel (or voxel) using the weighted sum of intensities within an adaptive 2D or 3D (or 2D or 3D + time) neighbourhood and to use image patches to take into account complex spatial interactions. The neighbourhood size is selected at each spatial or space-time position according to a bias-variance criterion. The algorithm requires no tuning of control parameters (already calibrated with statistical arguments) and no library of image patches. The method has been applied to real noisy images (old photographs, jpeg -coded images, videos, ...) and is exploited in different biomedical application domains (time-lapse fluorescence microscopy, video-microscopy, mri imagery, x -ray imagery, ultrasound imagery, ...).
The fast -2d -safir software (APP deposit number: IDDN.FR.001.190033.001.S.A.2007.000.21000) written in c ++ removes mixed Gaussian-Poisson noise in large 2D images, typically pixels, in a few seconds. The method is unsupervised and is a simplified version of the method related to the safir -nD software. The software dedicated to microarrays image denoising, was licensed to the INNOPSYS company which develops scanners for disease diagnosis and multiple applications (gene expression, genotyping, aCGH, ChIP-chip, microRNA, ...).
| On-line demo: Mobyle@GenOuest Bioinformatics http://mobyle.genouest.org/cgi-bin/Mobyle/portal.py#forms::NDSafir |
| Free download binaries: Binaries of the software nD-safir are freely and electronically distributed. Developed in standard C/C++ under Linux using the CImg library, it has been tested over several platforms such as Linux/Unix, Windows XP and Mac OS. |
| Academic licence agreements: Institut Curie, CNRS, ENS Ulm, Oxford university, Weizmann Institute, UCSF San-Francisco, Harvard university, Berkeley university, Stanford university, Princeton university, Georgia-Tech, Kyoto university, IMCB Singapore ... |
| Partners: J. Boulanger, J. Salamero (UMR 144 CNRS Institut Curie), P. Elbau (RICAM Linz, Austria), J.B. Sibarita (UMR 5091 University of Bordeaux 2). |
|
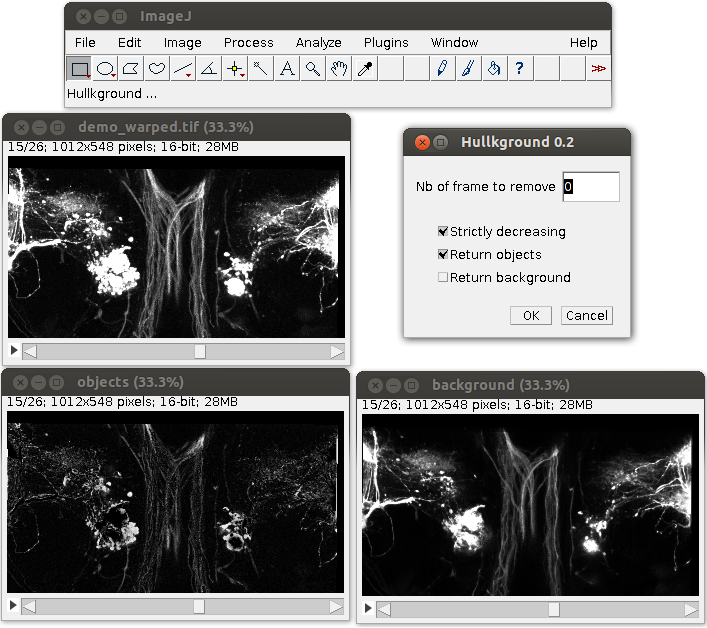
HullkGround: Background subtraction by convex hull estimation
The HullkGround software (APP deposit number: IDDN.FR.001.400005.000.S.P.2009.000.21000) written in java (plug-in ImageJ , see Fig. 3 ) decomposes a fluorescence microscopy image sequence into two dynamic components: i/ an image sequence showing mobile objects; ii/ an image sequence showing the slightly moving background. Each temporal signal of the sequence is processed individually and analyzed with computational geometry tools. The convex hull is estimated automatically for each pixel and subtracted to the original signal. The method is unsupervised, requires no parameter tuning and is a simplified version of the shapes-based scale-space method [25] .
On-line demo: Mobyle@GenOuest Bioinformatics http://mobyle.genouest.org/cgi-bin/Mobyle/portal.py#forms::Hullkground
Partners: A. Chessel and J. Salamero (UMR 144 CNRS Institut Curie)