Section: Software
Image Processing software distribution
Participants : Tristan Lecorgne, Charles Kervrann.
The objective is to disseminate the distribution of serpico image processing software for biologist users:
Free binaries: software packages have been compiled for the main operating systems (Linux, MacOS, Windows) using CMake (see http://www.cmake.org/ ). They are freely available on the team website under a proprietary license (e.g. nD-Safir and Hullkground are distributed this way at http://serpico.rennes.inria.fr/doku.php?id=software:index ).
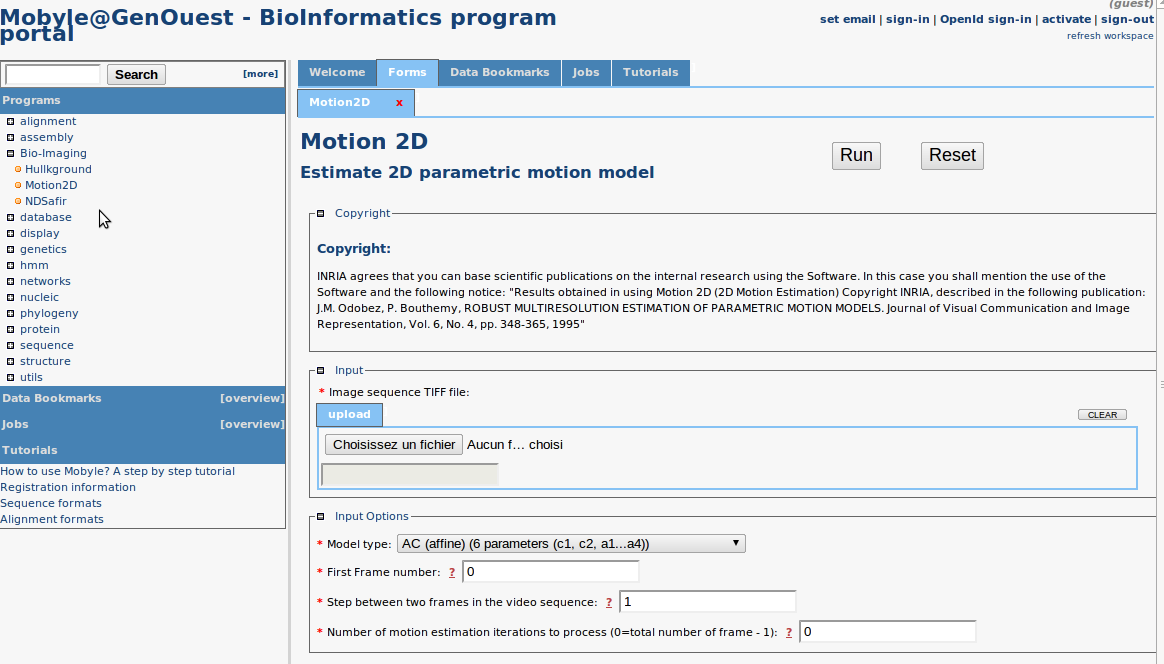
Mobyle web portal: An online version of the software has been released using the Mobyle framework (see http://mobyle.pasteur.fr/ ) developed at Institut Pasteur. The main role of this web portal is to demonstrate the performance of the programs developed by the team. The web interface makes our image processing methods available for biologist users without any installation or configuration (see nD-Safir , Hullkground , Motion2D (see Fig. 5 ) at http://mobyle.genouest.org/ ). The size of submitted images is limited by network bandwidth. We use the computing facility of the GenOuest platform to run calculations.
ImageJ plug-ins: ImageJ (see http://rsb.info.nih.gov/ij/ ) is a widely used image visualization and analysis software for biologist users. We have developed ImageJ plug-in java versions of the following software: nD-Safir , Hullkground (see Fig. 3 ), Motion2D , HotSpotDetection and OpticalFlow .
Institut Curie database: Institut Curie is currently acquiring a new database system to store mass of data. The database can be searched via meta-data and includes menu selections that enable to run remote processing. We have integrated nD-Safir in the interface environment to allow the database users to denoise images easily.
Partners: C. Deltel (Inria Rennes SED) and Perrine Paul-Gilloteux (UMR 144 PICT IBiSA CNRS Institut Curie)