Section: New Results
Robust parametric stabilization of moving cells
Participants : Solène Ozeré, Patrick Bouthemy, Charles Kervrann.
Paper under review.
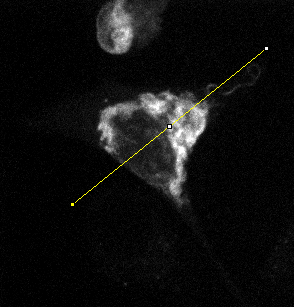
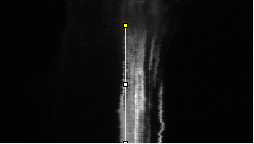
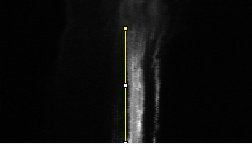
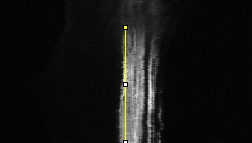
Analysing the dynamic behaviour of individual particles (e.g., proteins, vesicles) inside a cell is of primary importance in cell biology. However, the motion of these particles observed in live cell microscopy image sequences is the addition of the global movement of the cell and their own single motions. Hence, automatically stabilizing the cell (or a group of cells), i.e. compensating for its global motion or equivalently registering its successive positions over time, is previously required. We have proposed a cell stabilization method based on a realtime robust multiresolution scheme (Motion2D software [36] ). It can simultaneously handle the estimation of 2D parametric global motions (e.g., affine motion model) and of temporal intensity variations. Three temporal intensity models have been investigated: constant additive model, exponential decay model (corresponding to the photobleaching effect), continuity equation. We have carried out experiments on three biological situations: development of cells, displacements of endosomes, protein recruitment by the Golgi. We have demonstrated the accuracy of our method on these challenging examples and its capacity to efficiently reveal the own motion of subcellular particles. It yields better results than the StackReg method (http://bigwww.epfl.ch/thevenaz/stackreg/ ), classically used in the field, in cases involving strong local dynamics (see Fig. 6 ).
Partners:: Perrine Paul-Gilloteaux (UMR 144 CNRS PICT IBiSA Institut Curie)
|