Section: New Results
Image deconvolution algorithms for tagged-RNA and gene localization in live yeast
Participant : Charles Kervrann.
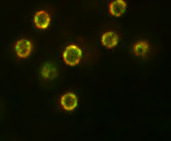
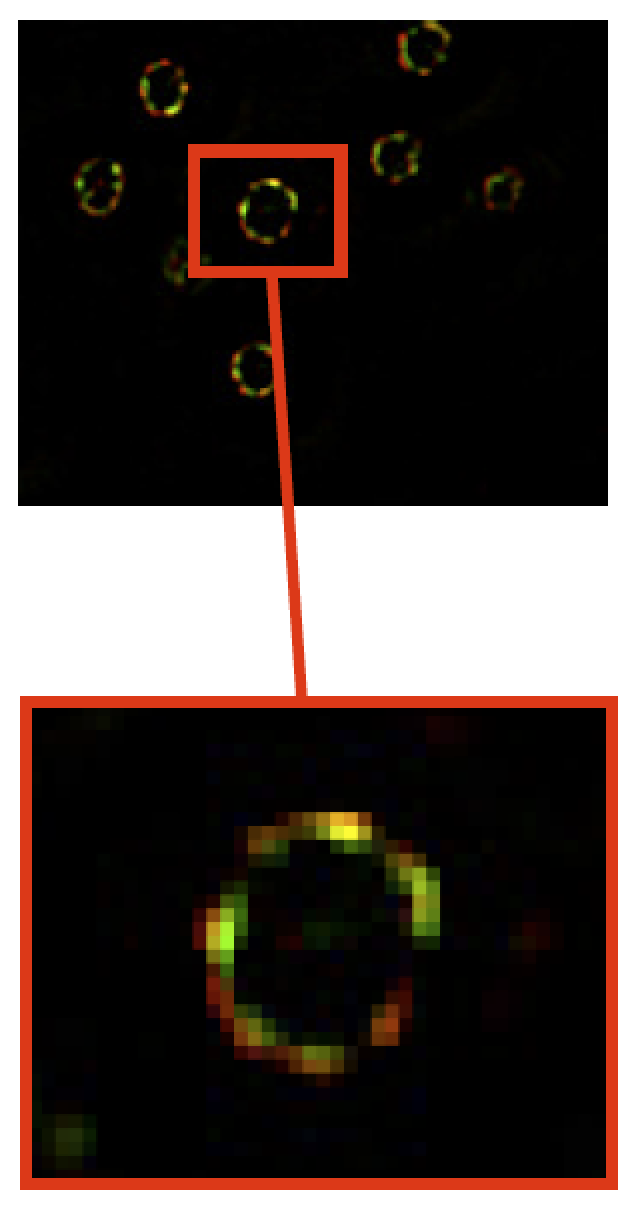
In fluorescence microscopy, the image quality is limited by out-of-focus blur and high noise. Traditionally, image deconvolution is needed to estimate a good quality version of the observed image. The result of deconvolution depends heavily on the choice of the regularization term. The regularization functional should be designed to remove noise while retaining the image structure. In this study, we investigated non quadratic regularization terms to preserve fine details of underlying structures and we studied appropriate optimization algorithms. The deconvolution method has been especially dedicated for 3D high-precision gene localization in cell nuclei [47] . For illustration, tagged gene (green marker) and tagged nucleoporins/nuclear periphery (red marker) are shown in Fig. 3 . A noisy and blurred image can affect the nuclear membrane estimation and gene detection and, consequently, the computed related distances.
Collaborators: Giovanni Petrazzuoli (Inserm U944, CNRS UMR 9212, Hôpital Saint-Louis, Paris),
Catherine Dargemont (Inserm U944, CNRS UMR 9212, Hôpital Saint-Louis, Paris),
Jean Salamero (UMR 144 CNRS-Institut Curie, PICT-IBiSA).