Section: New Results
Intracellular drift and diffusion coefficient estimation: a trajectory label-based approach
Participants : Antoine Salomon, Charles Kervrann.
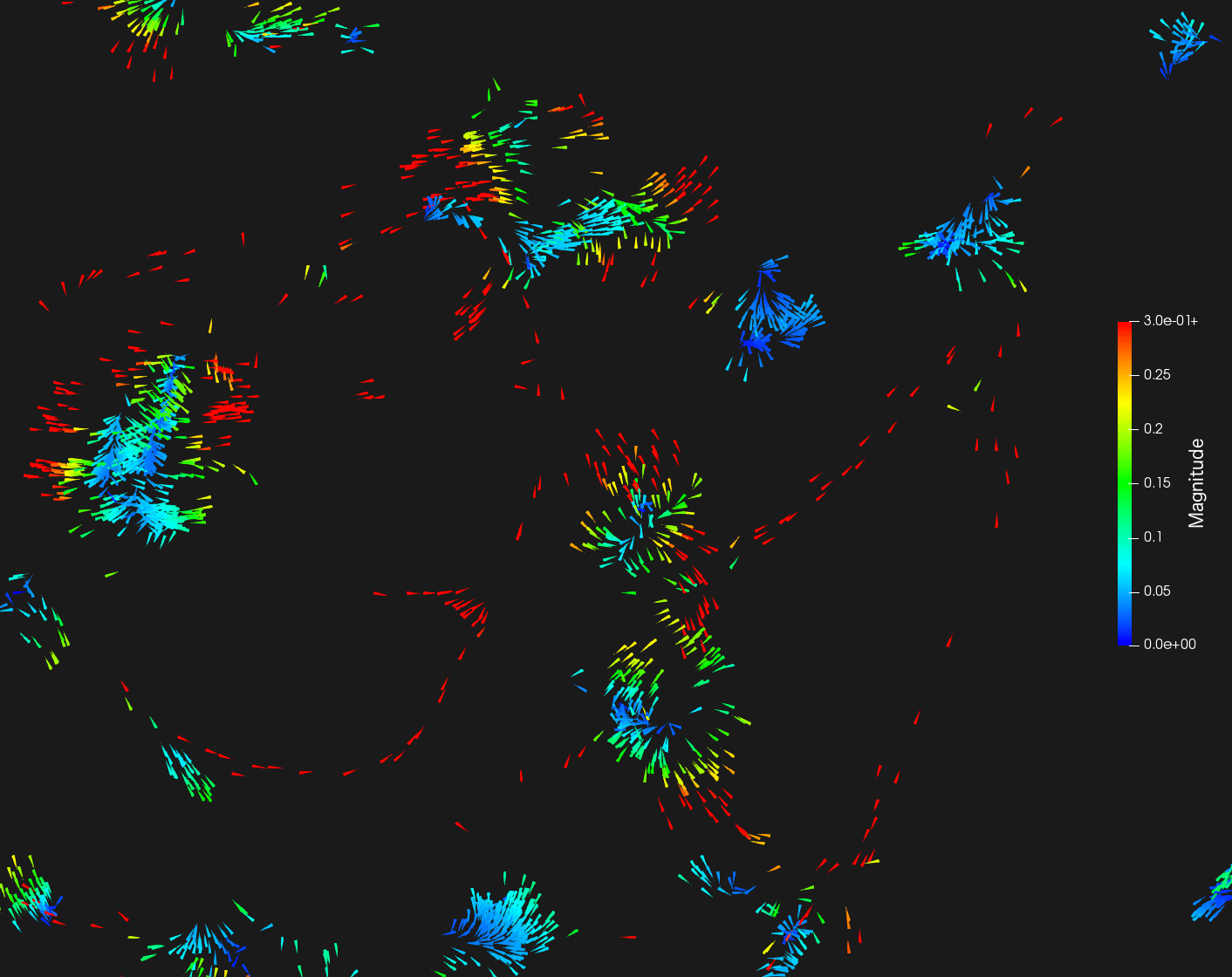
Dedicated computational methods for intracellular drift and diffusion map estimation rely on a scanning approach, using a sliding window (or cube, in the 3D case) in which all elementary particle movements taken from the trajectories inside the window/box are averaged [41]. This approach lacks precision, and a huge amount of trajectory data is needed to obtain significative results. In our new approach, we currently investigate several high-level features to obtain more satisfying results even from a small amount of data. First, we exploit classification of sub-trajectories (Brownian motion, superdiffusion, subdiffusion) obtained in the Lagrangian setting [16]. This classification is used to select trajectories of the same type in a local region and to guide weighting averaging. It allows us to calculate the drift separately on each type of movement, which avoids confusion between Brownian, confined and directed motions (see Fig. 6). Furthermore, the calculation is efficiently performed on trajectory-sliding kernels instead of scanning windows to save computing time in 2D and 3D. We considered square-box (sliding window), circle-box, cone-shaped, and Gaussian-shaped kernels.
Collaborators: Vincent Briane (UNSW Sydney, School of Medical Sciences, Australia),
Myriam Vimond (CREST ENSAI Rennes),
C.A. Valades Cruz and C. Wunder, (Institut Curie, PSL Research University, Cellular and
Chemical Biology, U1143 INSERM / UMR 3666 CNRS).